
Specifically SEQSEE is a multi-purpose menu-driven suite of Packages at the "cost" of the freely available shareware products. SEQuence SEEker) which offers the program diversity of the expensive
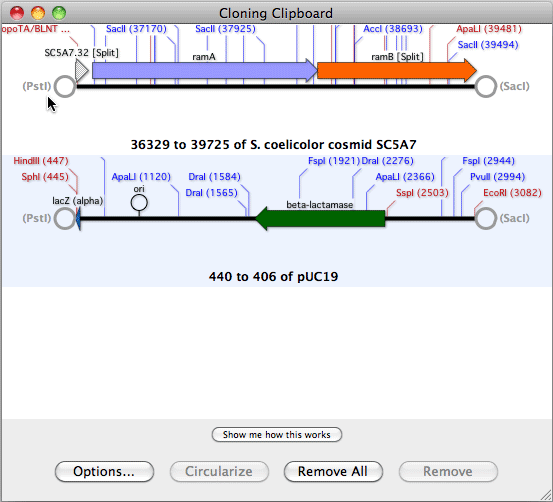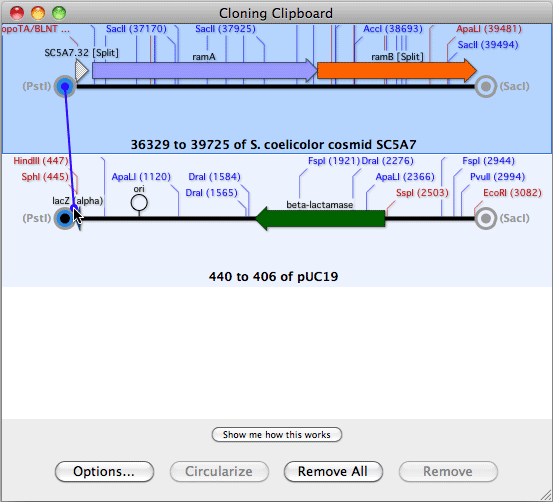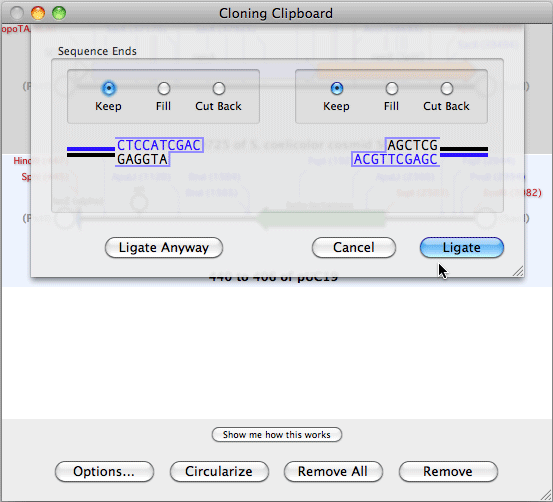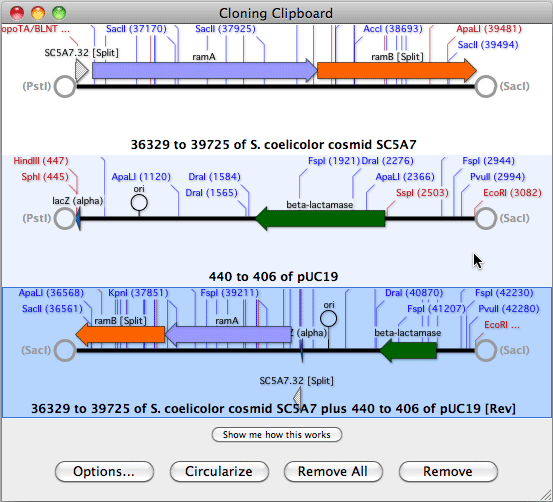
HJOW TO USE ALIGN TO REFERENCE IN MACVECTOR SOFTWARE
Program diversification and this software stratification we haveĮndeavored to develop a publicly available software package (called SEQSEE Products are becoming less and less useful. The field with the most powerful (and most useful) packages becoming moreĪnd more expensive while the freely available, unintegrated "shareware" Result, a rather problematic "software stratification" is developing in Of course with this rapid expansionĬomes the usual problems of limited availability, restricted usabilityĪnd increased costs of most sequence analysis software products. Into a phase of very rapid expansion with many new and unforeseenĪpplications being proposed for an increasingly diverse and substantially Quite clearly the development of sequence analysis packages is entering To permit even more diverse inquiries and analyses. New databases containing secondary structural information, phi and psiĪngles, torsion angle restraints, NMR chemical shifts, sequence motifsĪnd the like, are continually being added to the current software arsenal "homology" or "sequence similarity" (Gribskov et al., 1987). Likewise, the push toĭevelop more efficient methods for identifying the potential function orĪctive site location of newly isolated proteins is leading to theĭevelopment of methods which are, in effect, redefining the meaning of Previously uncharacterized proteins (Schulz, 1988). Prediction algorithms) being used to predict the tertiary structure of Programs (involving multiple sequence alignments and advanced structure Increasingly common to see some of the more advanced sequence analysis Important "computer experiments" has induced others, including X-rayĬrystallographers, NMR spectroscopists, protein engineers and evolutionaryīiologists to begin using or adapting these same software packages to helpĪnswer specific questions of their own. The success that molecular biologists have had at performing simple, yet Products (see Doolittle, 1990 and references therein).
Oncongenic products and the establishment of evolutionary relatednessīetween hundreds of previously unidentified or poorly understood protein Repetitive or recurrent folding "modules", the identification of various

New and important receptor families, the identification of numerous These include, just to mention a few, the identification of a number of Of extremely important and very useful "discoveries" have been made. Packages (in conjunction with their accompanying databases) that a number It is a result of the widespread implementation of these software Permit flexible sequence manipulations such as database searching,Īligning, comparing and matching - all at the touch of a button. Many of these larger and more costly programs Wisconsin's GCG suite, IBI's MacVector, and others are now widelyĪvailable for this purpose and are typically designed to run onĬomputers of all sizes and shapes including IBM PC's, Macintosh's, VAX's,Īnd SUN workstations. Programs or programming suites such as Intelligenetics' IG suite, Otherwise dissect sequence data in a useful or informative manner. Which would permit molecular biologists to quickly compare, analyze and As a consequence, a great deal of privately fundedĮffort has been directed towards the development of software (and hardware) Sequence information, it has not necessarily solved the problem of its While the development of theseĬentralized databases has certainly helped in the rapid dissemination of Sequence data is readily available, in computer readable format, to any Headache of keeping up with this information explosion. Which is being generated every single day.įortunately, through the establishment of publicly funded databanks suchĪs GENBANK, SWISS-PROT, NBRF-PIR and EMBL, most of us have been spared the Impossible to attempt to analyze or categorize this huge reservoir ofīiological information or to manage the rapid influx of new sequence data Without the aid of a computer it would be simply For example, since 1980 the number of sequences in the PIRĭatabank alone has increased from less than 2000 to almost 45,000 separateĮntries today. Protein and nucleic acid sequence data that has been generated in the lastĭecade. This is in no small part due the vast quantity of raw The past few years have seen an explosion in the use of computers in Contains a number of functions for analyzing protein sequences.


 0 kommentar(er)
0 kommentar(er)
